BOolean Operation based Screening and Testing (BOOST) is a method for detecting gene-gene interactions. It allows examining all pairwise interactions in genome-wide case-control studies in a remarkably fast manner. Interaction analyses on seven data sets from the Wellcome Trust Case Control Consortium were carried out. Each analysis took less than 60 hours to completely evaluate all pairs of roughly 360, 000 SNPs on a standard 3.0 GHz desktop with 4G memory running Windows XP system.
The definition of gene-gene interactions used in BOOST
BOOST uses widely accepted definition of gene-gene interactions which is based on logistic regression models. The logistic regression model with only main effects (i.e., the main effect model) has the following form:
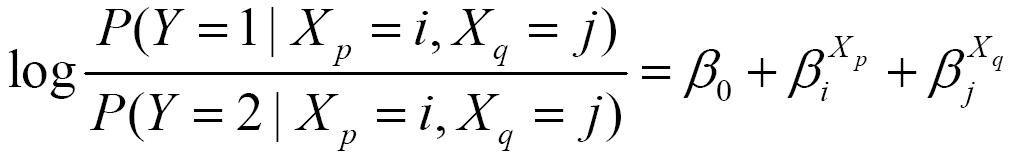
The logistic regression model with both main effect terms and interaction terms (i.e., the full model) has the following form:
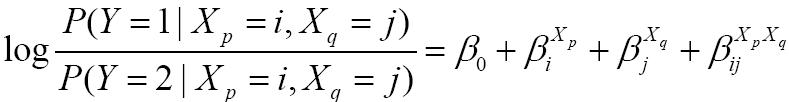
Let LM and LF be the maximum log-likelihoods of the main effect model and the full model, respectively. According to the likelihood ratio test, interaction effects are defined as the difference of their deviance, i.e., 2(LF-LM) .
Boolean operations
Based on the equivalence between a logistic regression model and its corresponding log-linear model, we construct our test statistic using log-linear models. The advantage of doing so is that we only need to fill the contingency tables. We design a Boolean representation of genotype data, which promotes the CPU efficiency because it only involves Boolean values and allows using fast logic (bitwise) operations to obtain the contingency tables.
Screening and Testing
In the space of log-linear models, the homogeneous association model is the equivalent form of the main effect logistic regression model, and the saturated model matches the full logistic regression model. Since no closed-form solution exists for maximum likelihood estimation of the homogeneous association model, we propose a two-stage (screening and testing) method. In the screening stage, we use a non-iterative method to approximate the likelihood ratio test statistic in evaluating all pairs of SNPs and select those passing a specified threshold. Most non-significant interactions will be filtered out. The survival of significant interactions is guaranteed becasue of the tight approximation bound. In the testing stage, we employ the classical likelihood ratio test to measure the interaction effects of selected SNP pairs.
The source, Win 32 and Win x64 executable file of BOOST is available : here (Downloads = ).
You may also download the files at a mirror site in http://snpboost.sourceforge.net.
For any enquiry, please contact YANG Can at macyang@ust.hk or Xiang Wan at xiangwan5@gmail.com.
Case 1 -- Disease loci with main effects: here.
Case 2 -- Disease loci without main effects: please see the reference Velez, D.R., White, B.C., Motsinger, A.A., Bush, W.S., Ritchie, M.D., Williams, S.M., and Moore, J.H. (2007). A balanced accuracy function for epistasis modeling in imbalanced datasets using multifactor dimensionality reduction. Genet. Epidemiol. 31, 306–315. .
17 - 09 - 2010 -- Source code of BOOSTw32 and Its executable version is added to BOOST.zip.
10 - 09 - 2010 -- BOOST web site is ready with BOOST's source and Win x64 executable.
"BOOST: a fast approach to detecting gene-gene interactions in genome-wide case-control studies",
The American Journal of Human Genetics. 2010. [pdf] [suppl]